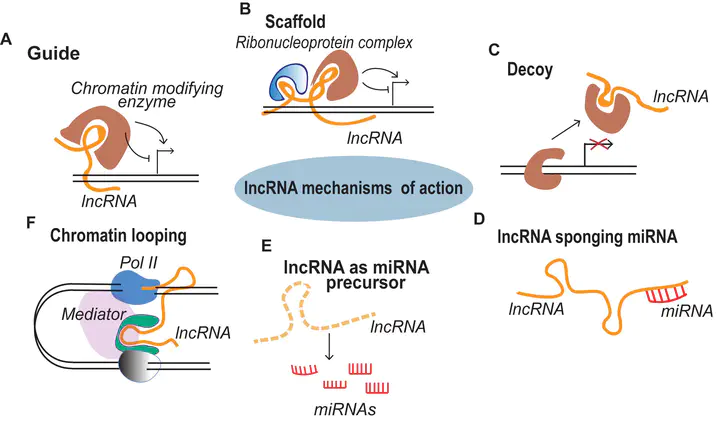
LncRNAs mode of action
Long non-coding RNAs (lncRNAs) are not as much studies as their protein coding counterpart. The specific mechanisms through which most of them act remain poorly understood. Therefore, only a very small number have been characterised for their molecular functions, both in animals and plants, and most of the time restricted to only a single species.
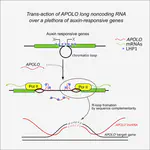
Using lines deregulated in the expression of selected lncRNAs, we can confirm their inferred targets. The expression levels of the predicted targets is correlated to the ones of the lncRNA. The regulation of transcript accumulation by lncRNAs, act mainly through chromatin conformation modification, epigenetic and chromatin mark modification and/or recruitment of transcription factors. Using the mis-expressed lncRNA lines, we check these different hypotheses. For example, how the deregulation of the lncRNA correlates with modifications of chromatin marks (H3K27me3, DNA methylation, …), chromatin conformation (chromatin accessibility and chromatin loops) and interactions with known lncRNAs partners. We already caracterised the action of some lncRNAs such as APOLO acting through R-loops that modulate local chromatin 3D conformation or MARS controling the formation of an ABA responsive chromatin loop betwwen an enhancer and its regulated gene MRN1.
Our investigations are not restricted to known mechanisms and we explore new ones. Indeed, much remains to be known about lncRNA action in plants. To this aim we investigate the relation between the lncRNAs and known regulators previously identified in these molecular mechanisms, notably through co-immuno-precipitation with either the regulator or the lncRNA as probe combined with either locus specific approaches or without any priors.